Power Analysis by Data Simulation in R - Part II
Part II - simulating groups and powering t-tests
The Power Analysis by simulation in R for really any design - Part II
This is Part II of my tutorial on how to do power-analysis by simulation. In Part I, we saw how to do a simulation for a simple toy-example with a coin-toss. In this part, we will use a more realistic problem that we might encounter in our daily research life and see how to simulate the power for these designs. By looking at how to do power-simulation for the independent-samples t-test and the paired t-test we will learn how to simulate normal-distributions, how to specify their effect-sizes, in terms of \(Cohen's\ d\). Moreover, we simulate correlated (i.e. multivariate) normal distributions in cases where we have correlated observations (e.g. paired-sample t-test). This will be an important tool for later parts of this tutorial.
In part III of this tutorial we will learn how we can conceptualize basically any design as a linear model and thereby be very flexible in our power analysis. In part IV we will learn how to apply this technique to complicated designs such as linear mixed-effects models and generalized mixed-effects models.
Simulating a between-subjects t-test
Let’s get to it.
The first thing we will need again in our simulation is one of the implemented simulation functions in R (those that let R run theoretical experiments for us), but this time it is not rbinom as we are not working with coin-flips but rnorm - the simulation function for the normal distribution.
Let’s have a short look at that function as we will keep working with it throughout the tutorial.
rnorm(n, mean, sd) takes three arguments, a sample-size n, a mean and a standard-deviation sd.
By specifying these values, we can sample random ‘people’ (or observations) that are participating in our simulated experiments.
Imagine, for example, that we have an intervention study in which we have a treatment group and a control group.
We can easily simulate both groups with rnorm but what should the means and sds of the groups be?
There are two ways we can approach this.
- We could think about what group means we expect in our given case and what we expect the spread of the groups to be on the measurment scale that we are working with. For example, if we use a 40-point scale for a clinical test we might know that a group with deficiencies on the thing that we measure would probably score around 10 points and that almost everyone from that group would score lower than 20 points. This statement (most people score around 10, almost everyone scores lower than tified as normal distribution with a mean of 10 and a standard-deviation of 5. In this case only 2.5% of the values (i.e. the values outside the 95% CI) will be higher than 20.
- In a new research project, we might not be able or willing to to make these statements. In this case, by making some extra assumptions, we can fall back to the approach that we also use in power-calculation software in most cases and define a standardized effect size that we can use to simulate data rather than defining the group means and standard-deviations directly.
I personally try to go with the first approach whenever possible, as I think that in many cases we know more about what we expect from our data than we think, even in new projects. Even if we do not know a lot about our data, we might still try out different assumptions (i.e. means and sds) for the groups in our simulation to see what power we would get for each of them. This way, we can make informed decisions about our sample size that are more nuanced than the one in which we just assume a standardized effect size and see what sample-size it implies and are forced to think harder about our data - something that might seem difficult and annoying at first, but is extremely useful and eduucational. Another advantage of specifying the groups directly is that we can do this for any arbitrarily complex design where standardized effect sizes are often difficult to calculate.
This said, for the cases where we might really not be willing to specify groups directly, and because it allows me to demonstrate some other interesting points, in this part I will discuss how we can use standardized effect-sizes in our simulation. In part III and IV however, we will always specify effects on the raw scale.
If we were using GPower now, we would most likely just fill in a difference between groups in \(Cohen's\ d\) and be done with it.
We could of course also follow this approac in a simulation by defining the groups based on the implied \(Cohen's\ d\).
For instance, we can just assume that group 1 as rnorm(n, 1,2).
Now, following from the formula for Cohen’s d:
\[Cohen's\ d = \frac{(M_1 - M_2)}{pooled \ sd}\]
where
\[pooled\ sd = \sqrt\frac{(sd_1^2+sd_2^2)}{2}\]
and adhering to the student t-test assumption of equal variances we can fill in the pooled sd formula above as
\[pooled\ sd = \sqrt\frac{(2^2+2^2)}{2} = 2\]
to get a \(Cohen's\ d\) of .50:
\[Cohen's\ d = \frac{(1 - 0)}{2} = 0.5\]
To get any other value for \(Cohen's\ d\) we can just change the pooled sd value to whatever we want. More generally, we want to solve the equation above for the pooled sd after specifying any \(Cohen's\ d\), e.g.:
\[0.5= \frac{(1 - 0)}{pooled\ sd}\]
We can solve an equation like that with R’s somewhat unintuitive solve function like this:
solve(0.5,1) # cohens d of .5## [1] 2solve(0.25,1) # cohens d of .25## [1] 4solve(2,1) # cohens d of 2## [1] 0.5giving us three examples of how we would need to specify pooled sd to arrive at a particular \(Cohen's\ d\).
Thus, if we want to do a t-test with two simulated groups and a cohen’s d of 0.5 we can simulate two groups of a particular sample-size by using the rnorm function.
Let’s say we have 30 participants in each group.
set.seed(1234)
group1 <- rnorm(30, 1, 2)
group2 <- rnorm(30, 0, 2)We can visualize the groups that we got in a plot like this:
hist(group1, col = "#addd8e", breaks = 10, main = "Histogram of both groups", xlab = "")
hist(group2, add = TRUE, breaks = 10, col= "#31a354")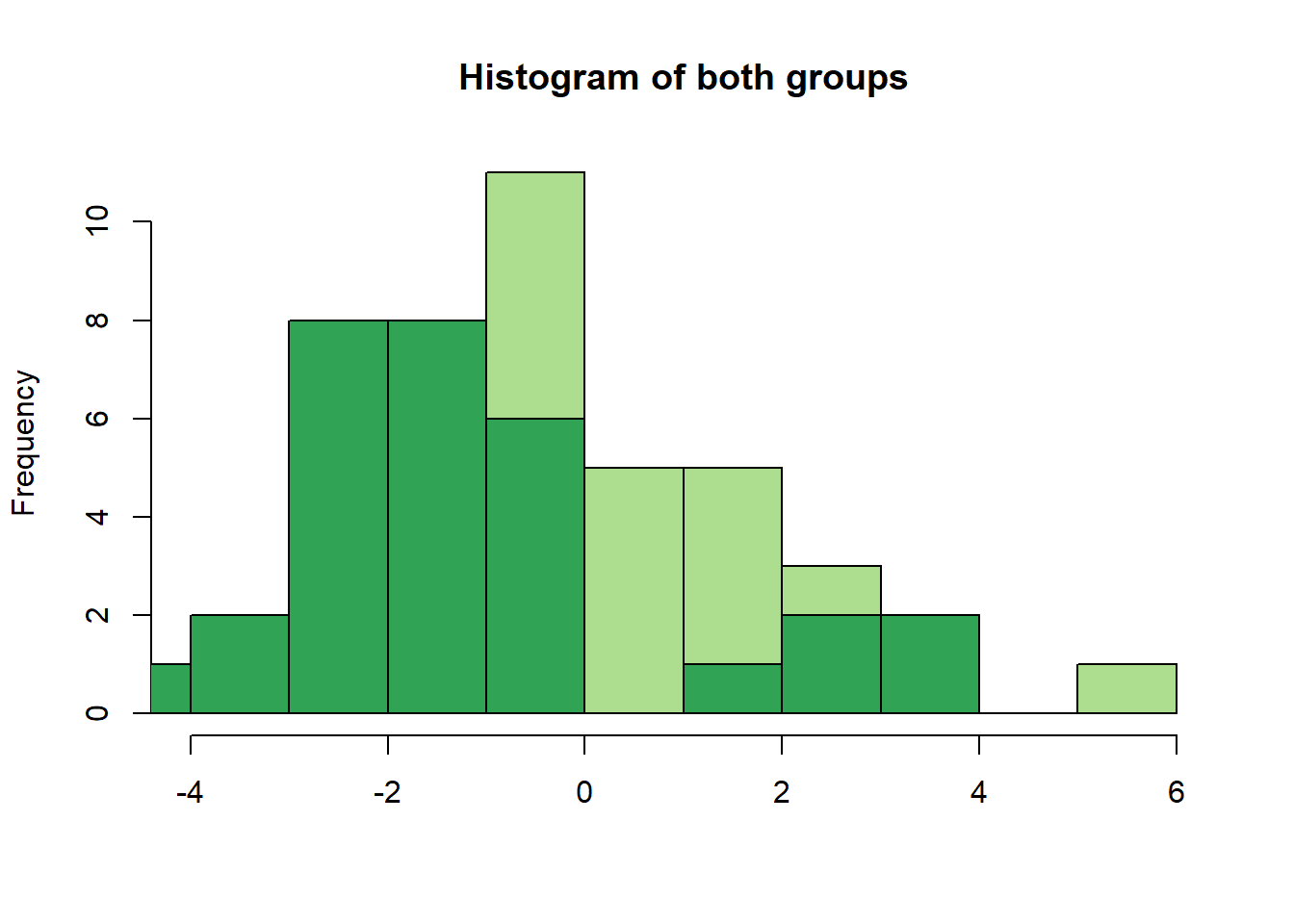
We can already make important observations from this plot:
We wanted to get normal distributions, but what we got here does not really look normal.
Why is that? Because we only have 30 people per group and taking only 30 values from the specified normal distributions does not really give us a good approximation of the real distribution.
This point is important: The sampling variability in such small groups is high and often, if small sample-studies (i.e. underpowered studies) find “effects”, they are often rather big and the consequence of this sampling variability rather than real differences of groups.
For example, by looking at the means of our sampled groups mean(group1) = 0.40715 and mean(group2) = -1.1032366 we see that the group mean of group 1 is actually closer to the mean that we specified for group 2 (i.e. 0) than to its own mean, while the mean for group 2 is far away from our intended mean.
Looking at the sds actually shows that they are quite close to what we wanted sd(group1) = 1.8059661 and sd(group2) = 1.9179992.
The \(Cohen's\ d\) that we wanted is also not presented very accurately at (mean(group1)-mean(group2))/(sqrt((sd(group1)^2+sd(group2)^2)/2)) = 0.8108043.
Again, if we would do this in Gpower, and specify a \(Cohen's\ d\), we will always work with an exact \(Cohen's\ d\), in a simulation approach we do not.
So let us run a t-test to see whether there is a significant difference here. First, we need to decide on an alpha-level again. What will we choose? Well, to have a good justification we have to elaborate on what the groups actually represent. Let us say that the difference between groups is related to an intervention that can elevate depressive symptoms. Thus, the control group (group1) did not get the intervention and scores higher on depressive symptoms while the treatment group (group2) is expected to score lower. Let us assume that this is the first study that we run and that, if we find anything we will follow it up by more extensive studies anyway. Therefore, we might not want to miss a possible effect by setting a too conservative alpha-level. If we find something in this study, we will conduct further studies in which we are more strict about the alpha level. Thus, we choose .10 for this first “pilot” study.
NOTE: The alpha-level “jusficications” in this tutorial are for educational purposes and to provide a starting point. They are obviously not as rigorous as we would like in a real research project. If you find yourself in a situation where you want to justify your alpha-level see Justify your alpha by Lakens et al. for a good discussion on this.
We can now run a t-test with R’s integrated t.test function.
t.test(group1, group2, paired = FALSE, var.equal = TRUE, conf.level = 0.9)##
## Two Sample t-test
##
## data: group1 and group2
## t = 3.1402, df = 58, p-value = 0.002656
## alternative hypothesis: true difference in means is not equal to 0
## 90 percent confidence interval:
## 0.7064042 2.3143690
## sample estimates:
## mean of x mean of y
## 0.407150 -1.103237The t-test shows, that this effect would be significant. However, we also got “lucky” and had a larger effect than we intended to have. To do a proper power analysis (lets say we first want to see whether 30 people per group are enough) we need to not only simulate each group once, but many many times and see how often we get a significant result at the desired alpha-level1. Moreover, we would like to have a power of at least 95%, again reflecting our view that we do not want to miss a possible effect.
In normal language these assumptions mean that if there is a difference, we will detect it in 19 out of 20 cases while, if there is no difference, we will only be incorrectly claiming that there is one in 1 out of 10 cases.
We will do this similarly to our simulations in part 1 of this tutorial.
set.seed(1)
n_sims <- 1000 # we want 1000 simulations
p_vals <- c()
for(i in 1:n_sims){
group1 <- rnorm(30,1,2) # simulate group 1
group2 <- rnorm(30,0,2) # simulate group 2
p_vals[i] <- t.test(group1, group2, paired = FALSE, var.equal = TRUE, conf.level = 0.90)$p.value # run t-test and extract the p-value
}
mean(p_vals < .10) # check power (i.e. proportion of p-values that are smaller than alpha-level of .10)## [1] 0.592Aha, so it appears that our power mean(p_vals < .10) = 0.592 is much lower than the 95% that we desired.
Thus, we did really get lucky in our example above when we found an effect of our intervention.
To actually do a legit power-analysis however, we would like to know how many people we do need for a power of 95 percent. Again we can modify the code above to take this into account.
set.seed(1)
n_sims <- 1000 # we want 1000 simulations
p_vals <- c()
power_at_n <- c(0) # this vector will contain the power for each sample-size (it needs the initial 0 for the while-loop to work)
cohens_ds <- c()
cohens_ds_at_n <- c()
n <- 30 # sample-size
i <- 2
while(power_at_n[i-1] < .95){
for(sim in 1:n_sims){
group1 <- rnorm(n,1,2) # simulate group 1
group2 <- rnorm(n,0,2) # simulate group 2
p_vals[sim] <- t.test(group1, group2, paired = FALSE, var.equal = TRUE, conf.level = 0.9)$p.value # run t-test and extract the p-value
cohens_ds[sim] <- abs((mean(group1)-mean(group2))/(sqrt((sd(group1)^2+sd(group2)^2)/2))) # we also save the cohens ds that we observed in each simulation
}
power_at_n[i] <- mean(p_vals < .10) # check power (i.e. proportion of p-values that are smaller than alpha-level of .10)
cohens_ds_at_n[i] <- mean(cohens_ds) # calculate means of cohens ds for each sample-size
n <- n+1 # increase sample-size by 1
i <- i+1 # increase index of the while-loop by 1 to save power and cohens d to vector
}
power_at_n <- power_at_n[-1] # delete first 0 from the vector
cohens_ds_at_n <- cohens_ds_at_n[-1] # delete first NA from the vectorThe loop stopped at a sample-size of n-1 = 84 participants per group.
Thus make a conclusion about the effectiveness of our intervention at the specified alpha-level with the desired power we need 168 people in total.
To visualize the power we can plot it again, just as in the first part of the tutorial.
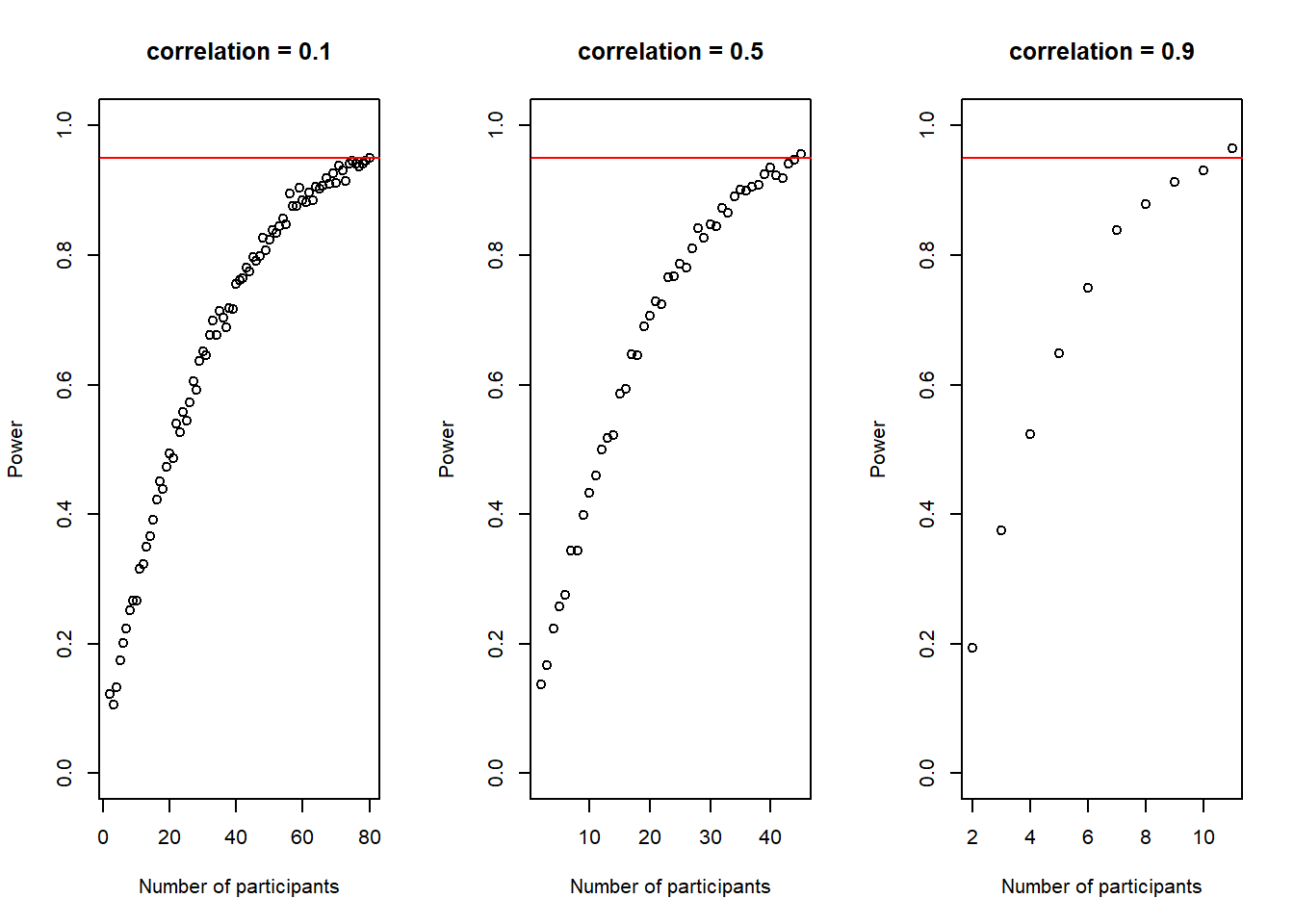
plot(30:(n-1), power_at_n, xlab = "Number of participants per group", ylab = "Power", ylim = c(0,1), axes = TRUE)
abline(h = .95, col = "red")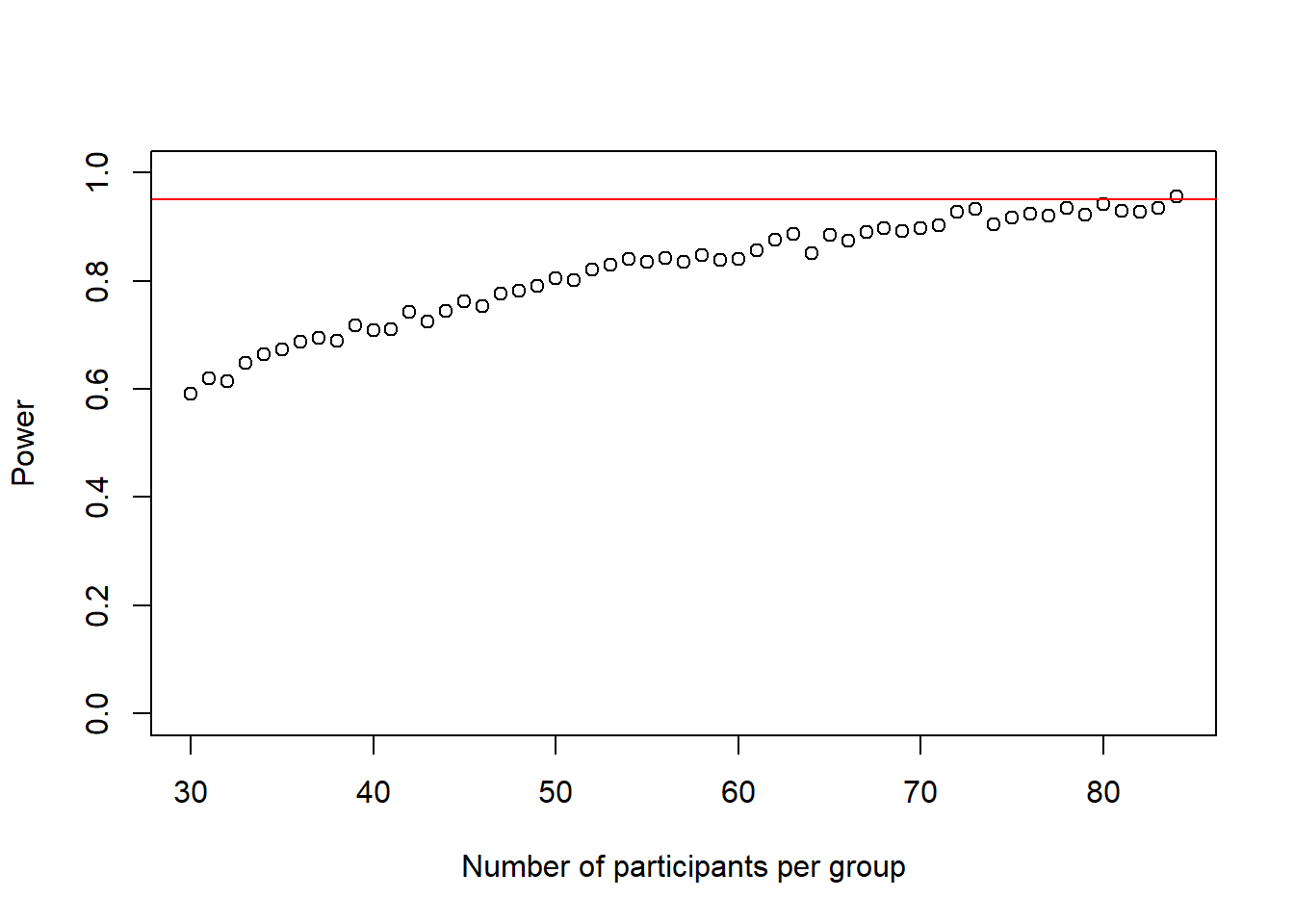
Again, this plot shows us how our power to detect the effect slowly increases if we increase the sample-size until it reaches our desired power.
There is another interesting observation to make here. In the code above, I also calculate the average \(Cohen's\ d\) for each sample size and the plot below shows how it changes with increasing sample-size.
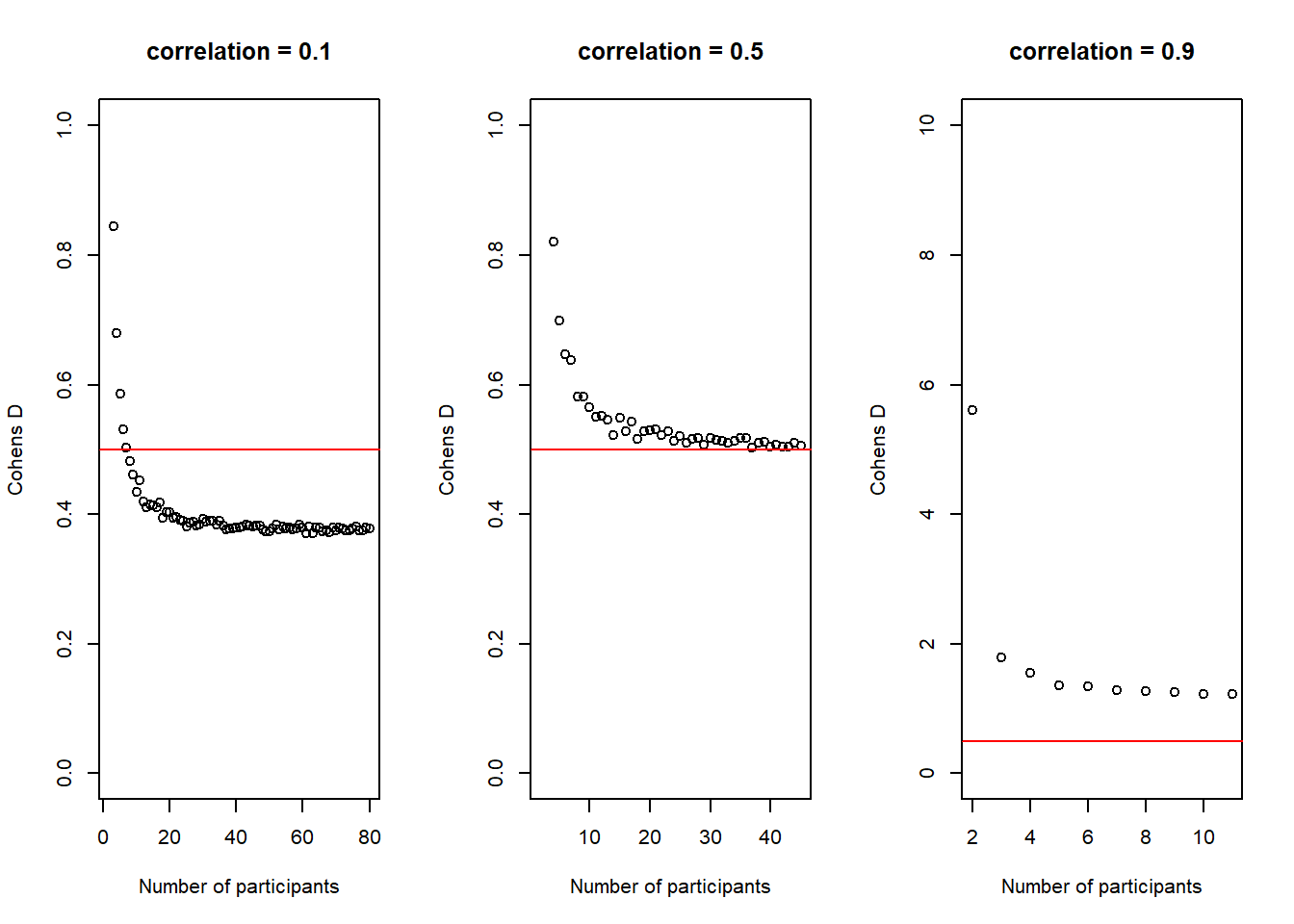
plot(30:(n-1), cohens_ds_at_n, xlab = "Number of participants per group", ylab = "Cohens D", ylim = c(0.45,0.55), axes = TRUE)
abline(h = .50, col = "red")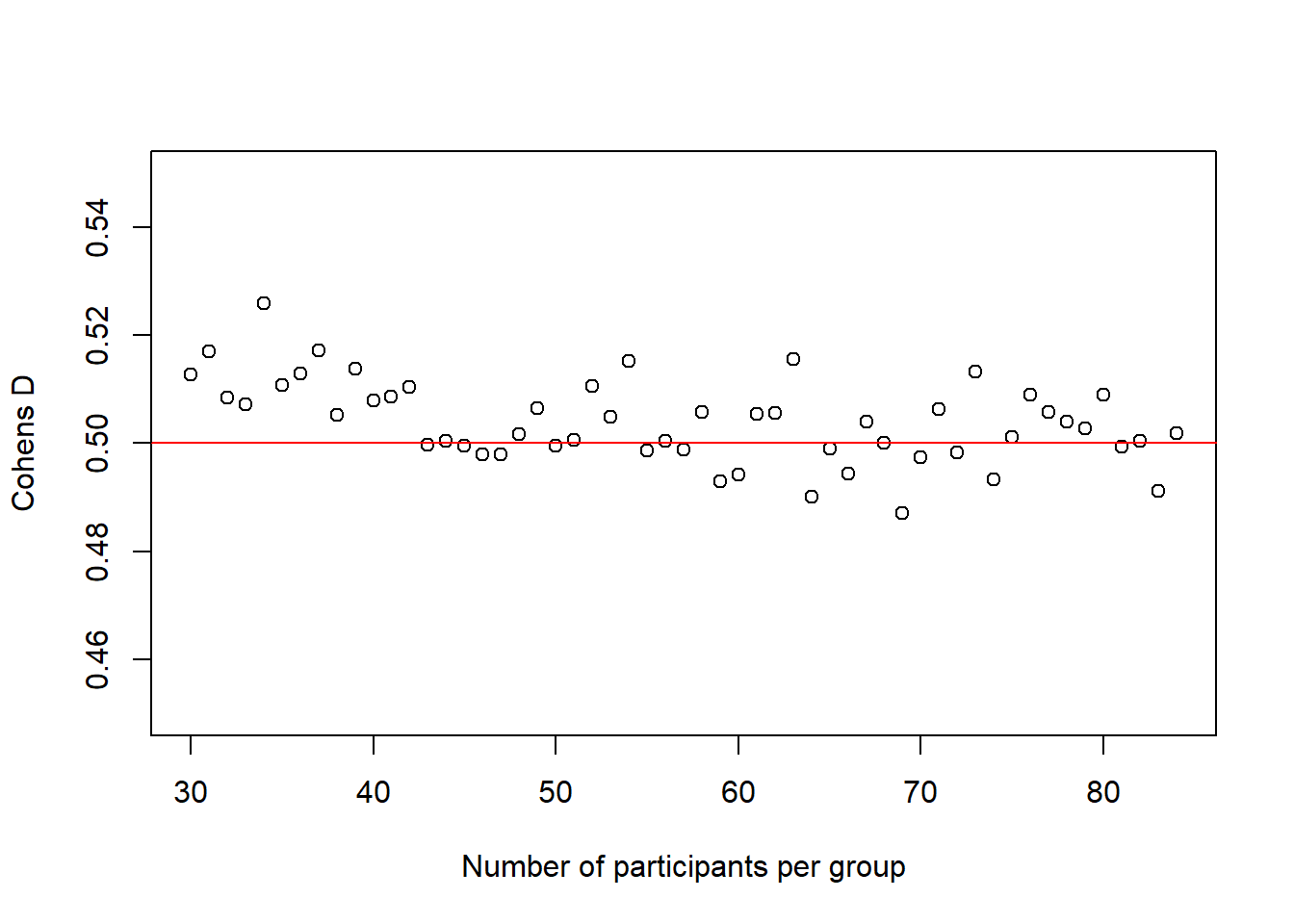
It is not super obvious in this plot and I had to change the scale of the y-axis quite a bit to make it visible, but we can actually see how our average \(Cohen's\ d\) initially deviates slightly more from the desired \(Cohen's\ d\) of .50 than in de end. In other words, in the beginning, for small sample-sizes there is more fluctuation than for bigger sample-sizes. That is pretty neat, as it seems very desirable that a power-estimation procedure takes into account that for smaller sample-sizes, even if the effect in the population is exactly the same (i.e. we always sample groups with a difference of \(Cohen's\ d\) = .50) it is just less precise.
Let’s have a brief summary of what we did so far. We just used the formula for \(Cohen's\ d\) to give our groups a certain difference that we are interested in, ran 1000 simulated experiments for each sample-size and calculated the power, just as in the first part of the tutorial.
However, I want to mention again that, even though it is convenient to specify the effect-size this way as it saves us from having to specify precise group means and standard-deviations directy and makes the specification more comparable, it is often preferable to specify the parameters on the original scale that we are interested in. This is especially the case if we have previous data on a research topic that we can make use of. Moreover, for more complex designs with many parameters, standardized effect sizes are often difficult to obtain and we are forced to make our assumptions on the original scale of the data. We will see this in later examples.
Simulating a within-subject t-test
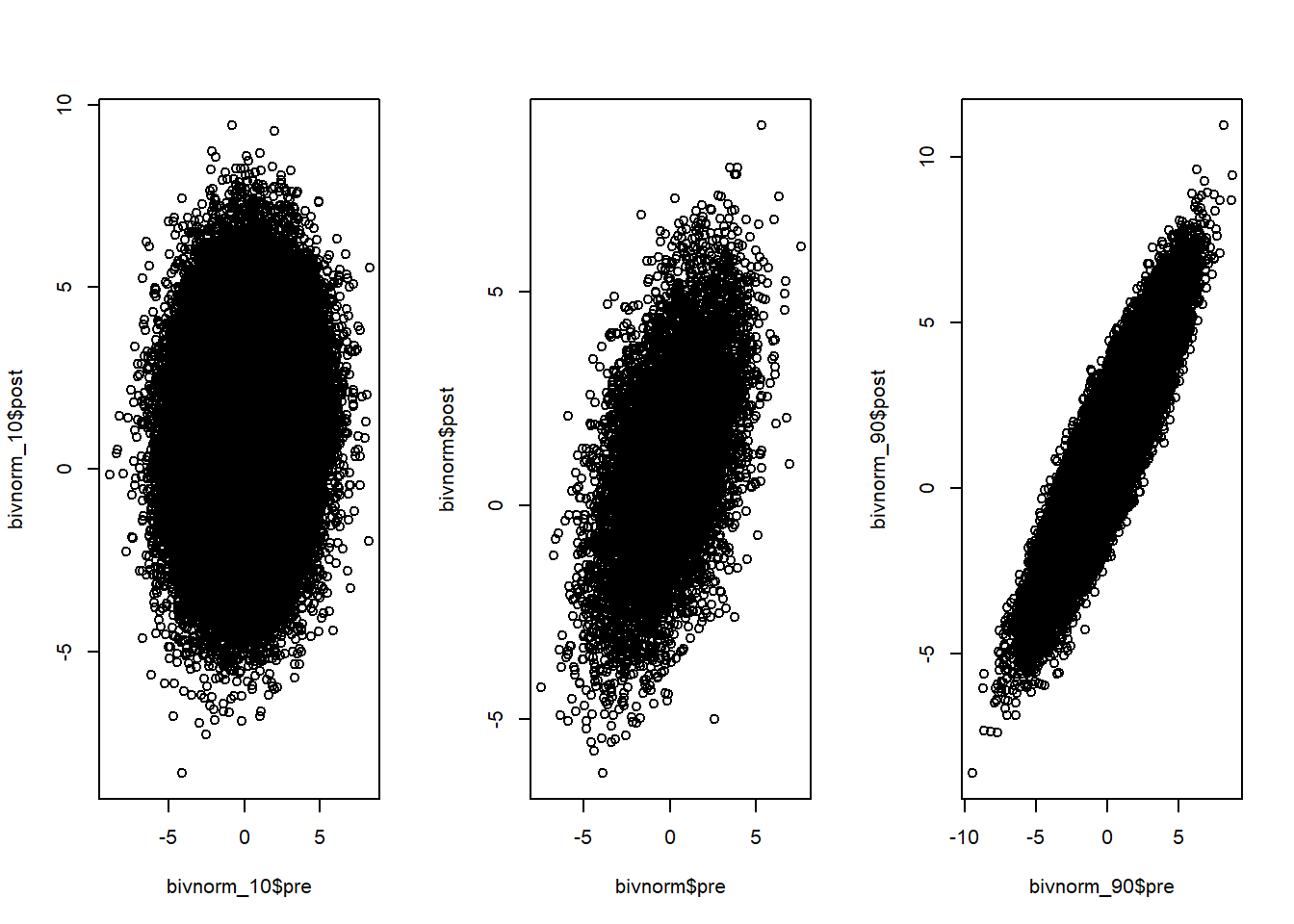
Intuitively, it might seem that we can use the exact same approach above for a paired t-test as well. However, the problem with this is that in a paired t-test we get 2 data-points from the same individual. For example, image we have a group of people that get an intervention and we measure their score before and after the intervention and want to compare them with a paired t-test. In this case, the score of the post-measure of a given individual is not completely independent of the score of the pre-measure. In other words, somebody who scores very low on the pre-measure will most likely not score very high on the post-measure and vice versa.
Thus, there is a correlation between the pre- and the post-measures in that the pre-measures already tell us a little bit about what we can expect on the post-measure. You probably already knew this but why does this matter for power simulation, you might wonder. It matters as it directly influences our power to detect an effect as we will see later. For now let’s just keep in mind that it is important.
So what do we do in a situation with correlated data as in the pre-post intervention situation? There are two ways we can go from here. First, we can simulate correlated normal distributions, as already mentioned above. However, for the particular case of a paired sample t-test, we can also just make use of the fact that, in the end, we are testing whether the difference between post- and pre-measures is different from 0. In this case, the correlation between the pre and the post-measure is implicitely handled when substracting the two measures. This way, we do not need to directly specify it. If the correlation is close to one, the standard-deviation of the difference scores will be very small, if it is zero, we will end up with the same situation that we have in the independent-sample t-test. Thus, we can just make use of a one-sample in which we test whether the distribution of difference-scores differs from zero as the paired t-test is equivalent to the one-sample t-test on difference scores (see Lakens, 2013 for more details on this).
Though the one-sample approach is easier to simulate, I will describe both approaches in the following as the first approach (simulating correlated normal-distributions) is more flexible and we need it for the situations we deal with later.
Using a one-sample t-test approach
When we want to do our power-calculation based on the one-sample t-test approach, we only have to specify a single difference-score distribution. We can do this again, based on the \(Cohen's\ d\) formula, this time for a one-sample scenario:
\[ Cohen's\ d = \frac{M_{diff} - \mu_0}{SD_{diff}}\]
In the above formula, to get our values for the simulation we can substitute the \(\mu_0\) by 0 (as our null-hypothesis is no difference) and solve the equation in the same way as above by fixing the mean-difference between pre- and post-measure, \(M_{diff}\) to 1 and calculating the sd we need for each given \(Cohen's\ d\), for instance
\[ 0.5 = \frac{1}{SD_{diff}}\]
putting this into Rs solve function again, we unsurprisingly get a 2 in this case.
solve(0.5, 1)## [1] 2To run our simulation we just need to modify the code above to run a one-sample t-test rather than a two-sample t-test and change the formula for \(Cohen's\ d\)
set.seed(1)
n_sims <- 1000 # we want 1000 simulations
p_vals <- c()
power_at_n <- c(0) # this vector will contain the power for each sample-size (it needs the initial 0 for the while-loop to work)
cohens_ds <- c()
cohens_ds_at_n <- c()
n <- 2 # sample-size
i <- 2
while(power_at_n[i-1] < .95){
for(sim in 1:n_sims){
difference <- rnorm(n,1,2) # simulate the difference score distribution
p_vals[sim] <- t.test(difference, mu = 0, conf.level = 0.90)$p.value # run t-test and extract the p-value
cohens_ds[sim] <- mean(difference)/sd(difference) # we also save the cohens ds that we observed in each simulation
}
power_at_n[i] <- mean(p_vals < .10) # check power (i.e. proportion of p-values that are smaller than alpha-level of .10)
cohens_ds_at_n[i] <- mean(cohens_ds) # calculate means of cohens ds for each sample-size
n <- n+1 # increase sample-size by 1
i <- i+1 # increase index of the while-loop by 1 to save power and cohens d to vector
}
power_at_n <- power_at_n[-1] # delete first 0 from the vector
cohens_ds_at_n <- cohens_ds_at_n[-1] # delete first NA from the vectorWe see that the loop stopped at n = 43 so the sample size we need is n-1 = 42
We can plot the power-curve again
plot(2:(n-1), power_at_n, xlab = "Number of participants per group", ylab = "Power", ylim = c(0,1), axes = TRUE)
abline(h = .95, col = "red")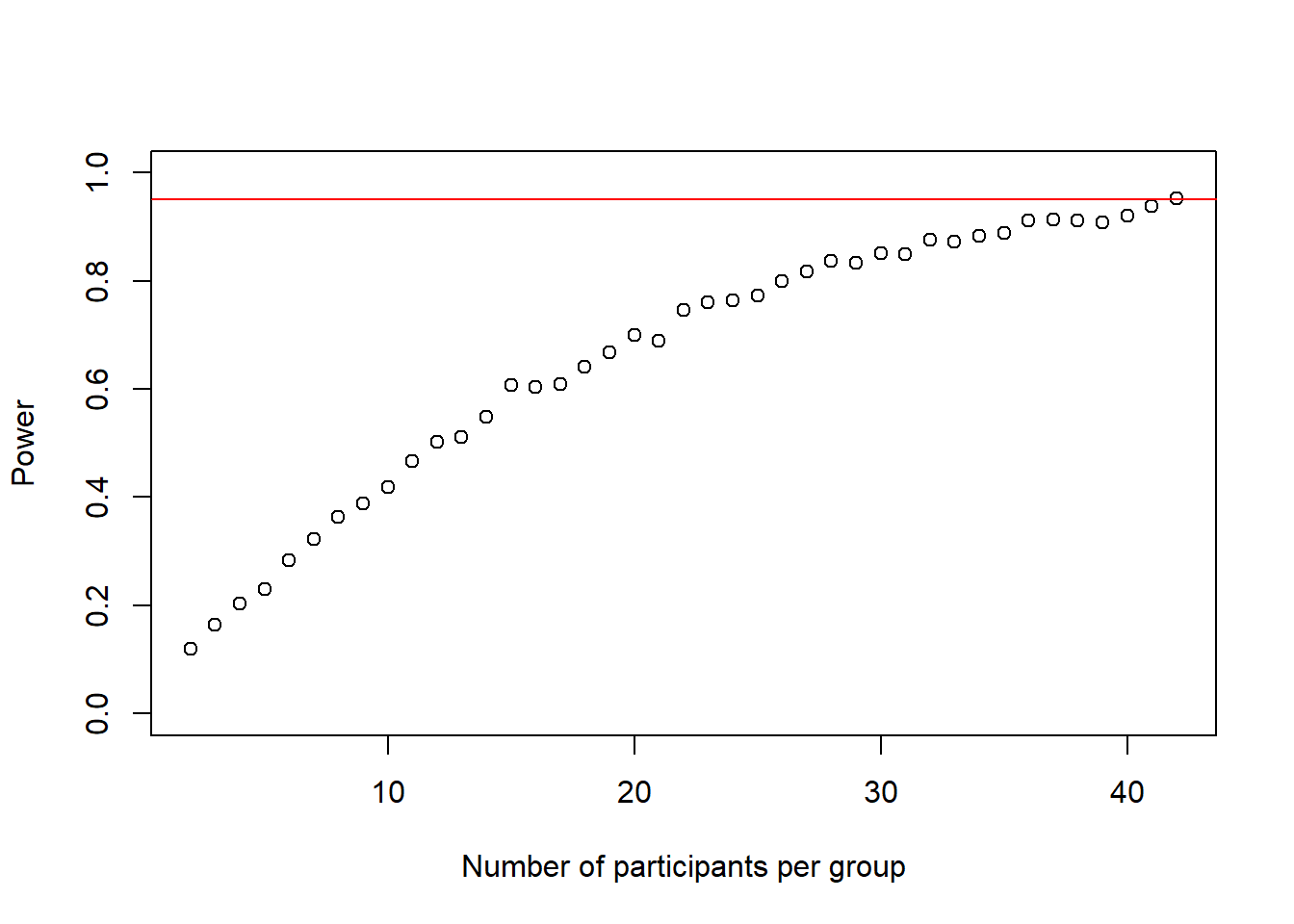
and the \(Cohen's\ d\) values:
plot(2:(n-1), cohens_ds_at_n, xlab = "Number of participants per group", ylab = "Cohens D", ylim = c(0.0,1.0), axes = TRUE)
abline(h = .50, col = "red")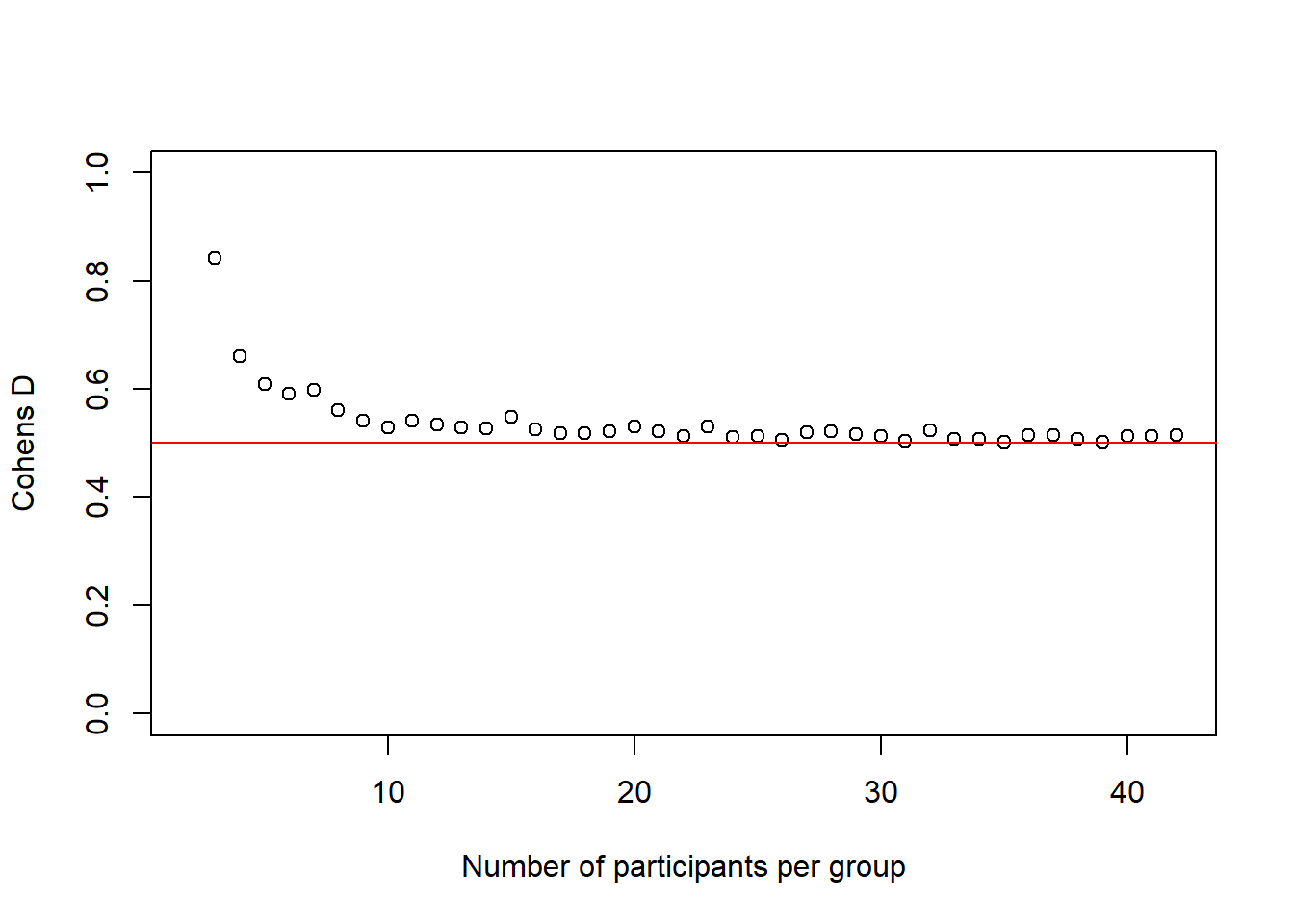
We see again, and this time more dramatically, how our simulated effect size becomes more accurate the bigger our sample gets.
Summary: Our first simulations with t-tests
This was the last bit that I wanted to discuss about simulating t-tests and the end of part II of this tutorial.
We have now learned how to simulate a t-test by using either \(Cohen's\ d\) as an effect-size estimate and, if necessary, tell R that our two groups, or measurements, are correlated in some way.
What we learned above is not restricted to doing t-tests however.
Simulating univariate (i.e. uncorrelated) or multivariate (i.e. correlated) normal-distributions will be what we do most of the time in part III and part IV of the tutorial.
The only thing that will change for more complicated designs is how we combine the different tools that we learned in this part to achieve our goal.
In part III of this tutorial, we will see how we can basically run every analysis as a linear model using the lm function instead of using the t.test function for t-tests, the aov function for ANOVA-designs and so forth.
By exploring how this works for t-test, anova and regression we will simulate our way through the third part and be flexible enough to simulate any classical research designs that we would, for example, be able to do in GPower. In part IV we will go beyond this and simulate mixed-effect models.
Footnotes
Think back to the possible sequences of coin tosses in part I. Instead of possible sequences of coin-tosses, we deal with possible sequences of people-scores here, assuming that they come from the underlying distribution that we specify. To get a good approximation of all the possible samples that we could get that still follow the specified distribution, we need to simulate many, many times.↩︎
This is how we solve for r:↩︎